MxIF Data in FUSION
How to load a new multiplexed immunofluorescence dataset locally in FUSION
Multiplexed Immunofluorescence Data (MxIF)
This diverse data type includes a variety of technologies which capture intensity of fluorescent probes across a section of tissue. Some examples include PhenoCycler, Cell-DIVE, and others.
Compared to spatial transcriptomics methods, MxIF methods are able to achieve single-pixel resolution for a smaller number of targets. This makes these types of techniques especially suited for low-informational, high-spatial granularity tasks such as simple cell annotation. By employing dimensional reduction techniques, several groups have demonstrated the ability of MxIF technologies to greatly expedite the process of cell annotation.
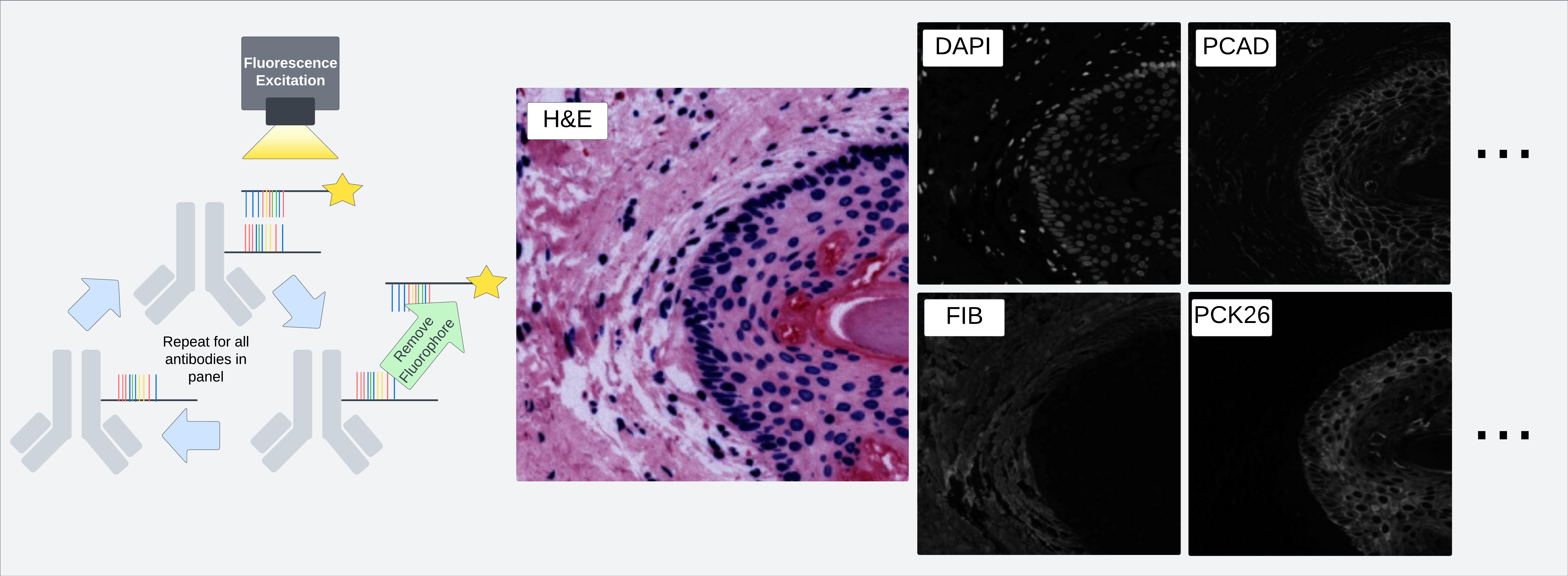 Figure demonstrating one process through which multiple fluorescent probes may be applied to a single section of an image to produce pixel-aligned images of multiple stains.
Figure demonstrating one process through which multiple fluorescent probes may be applied to a single section of an image to produce pixel-aligned images of multiple stains.
Checking Image Format Compatibility in large-image
Comparatively, MxIF datasets are some of the easiest to load into FUSION because they do not inherently include any annotations. To simply view an image consisting of several fluorescence channels, first check that your image type can be loaded in the default version of large-image installed along with fusion-tools:
1
2
3
4
5
6
7
8
9
10
11
import large_image
import json
# Specify the path to your image file
path_to_image_file = '/example/path/to/image.tiff'
# Using the .open method to create a "tile source" object
loaded_image = large_image.open(path_to_image_file)
# Printing large-image metadata for that file
print(json.dumps(loaded_image.getMetadata(),indent=4))
If you do not encounter any errors from that line, then that means that the image format your image file is in is included in the available formats for the default large-image installed with fusion-tools. If you do get an error, it’s likely you can simply add on one of the many available tile readers available through large-image. See the following webpage for more details: Image Formats. To install an additional tile source reader, you can use the following pip command with your desired tile source from the previous webpage.
1
pip install large-image[bioformats]
This example installs the “bioformats” tile-source reader.
The following line can be executed in Python to check your current installations list of supported sources:
1
2
3
4
5
6
7
8
import large_image
# Printing the image sources which are supported by your current installation
print(large_image.listSources())
# Printing the "priority" or the order in which different tile source readers will be attempted.
print(large_image.constants.SourcePriority)
Note If you know that your image should be multi-frame and the result of loaded_image.getMetadata() does not show multiple frames or the “frame” key, you will probably have to change which large-image sources you have installed. For example, the openslide reader does not support multi-frame images but is usually high on the priority list. Run the following command to uninstall specific tile source readers:
1
python -m pip uninstall large-image-source-openslide
Which will uninstall the openslide tile source reader
Loading MxIF Data in FUSION
Now that you have checked that your image is in a format that is readable by your environment’s large-image version, you can now create a layout for interactive analysis.
Some of the key components with MxIF datasets include:
MultiFrameSlideMap,HybridSlideMap,LargeMultiFrameSlideMap,LargeHybridSlideMap- These
SlideMaptypes are optimized for images consisting of multiple “frames” or fluorescence channels. Note: If you plan on analyzing both multi-frame and single frame (RGB) images in the same visualization session, use a “Hybrid” version. If you are loading cell annotations (>100k), it would be best to use one of the “Large” maps and setting themin_zoomargument to something manageable (like 4). This will greatly improve performance rendering annotations.
- These
ChannelMixer- This component controls which channels you see at the same time in your
MultiFrameSlideMaporHybridSlideMap. Select a channel and color for that channel and click the “Overlay Channels” button to view the assembled channels!
- This component controls which channels you see at the same time in your
Here’s an example layout:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
from fusion_tools.visualization import Visualization
from fusion_tools.components import HybridSlideMap, ChannelMixer
# Creating visualization object with one locally stored image and one row with two columns of components
vis = Visualization(
local_slides = ["/path/to/locally/stored/slide.tiff"],
components = [
[
HybridSlideMap(),
ChannelMixer()
]
]
)
vis.start()
Which will produce the following layout:
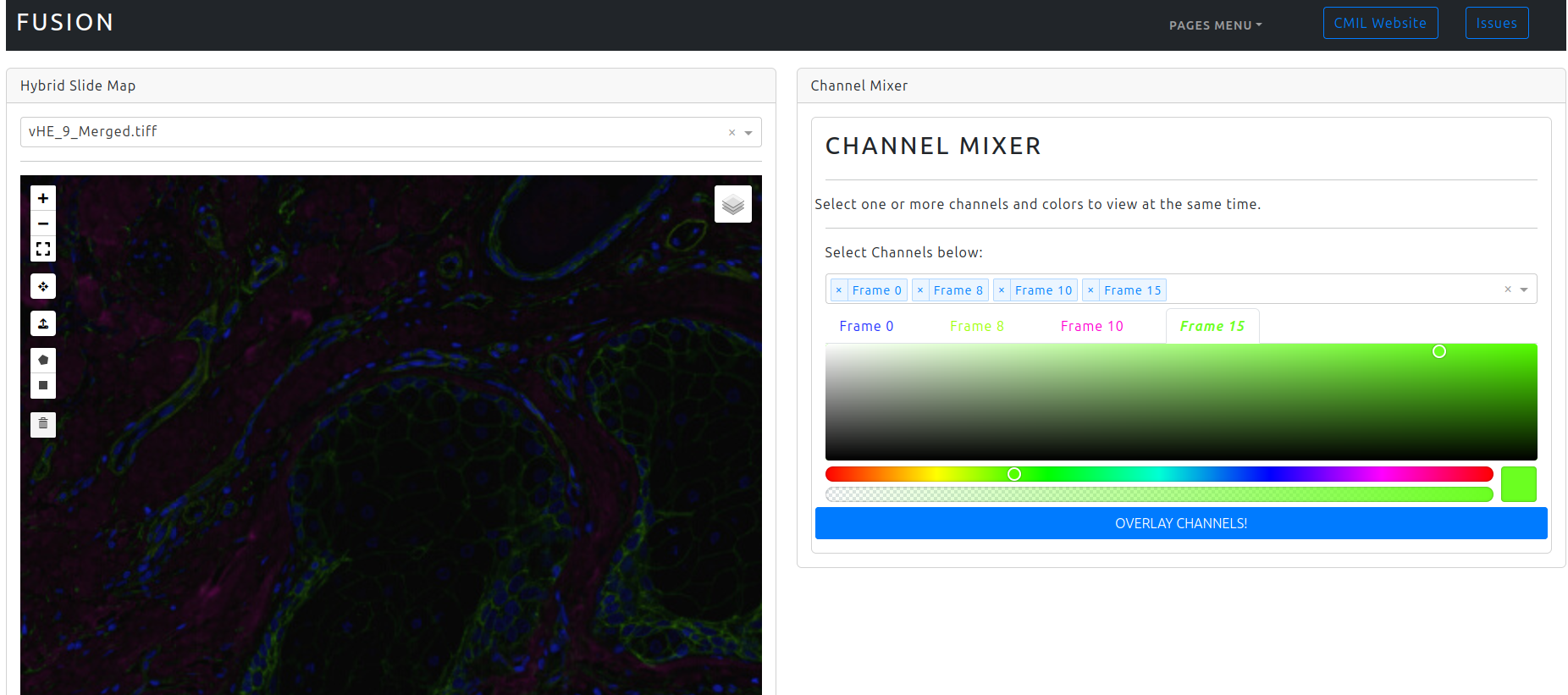 Cell-DIVE image from a skin sample with several overlaid frames.
Cell-DIVE image from a skin sample with several overlaid frames.