Welcome to FUSION
Background and motivation of FUSION
Background
Spatial –Omics Data
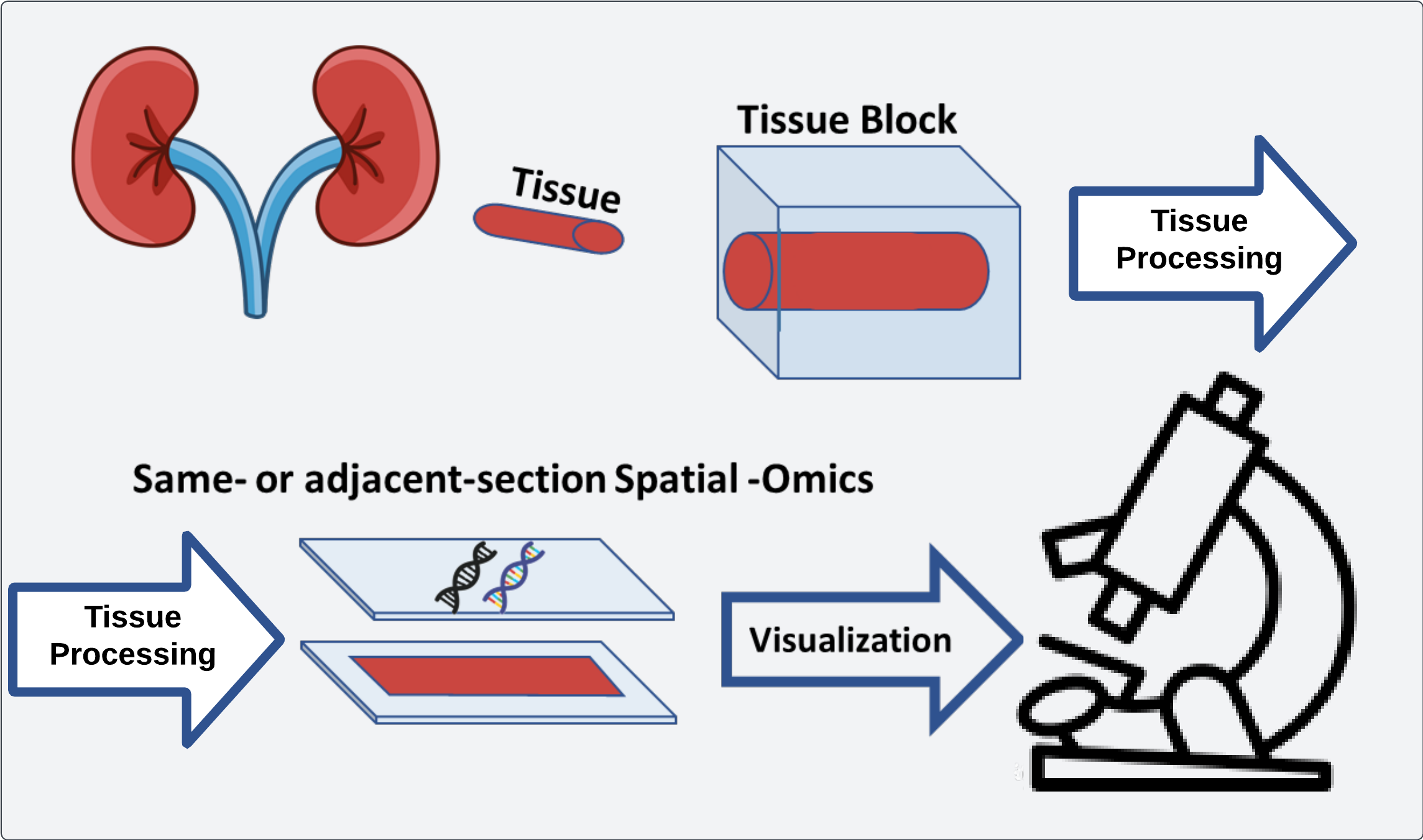 Workflow of spatial omics data where histology data is aligned with extracted omics data
Workflow of spatial omics data where histology data is aligned with extracted omics data
Spatial –omics data greatly expands the ability of researchers to pose complex hypotheses surrounding the spatial distribution of genomic markers. Compared to bulk sequencing, spatial –omics techniques focus on the preservation of where a particular signal is derived from relative to other signals in the same experiment. This data allows for testing colocalization of certain –omics (transcriptomics, metabolomics, proteomics, etc.) with other characteristics of a dataset as well as relational properties to other areas of a sample.
Histopathology
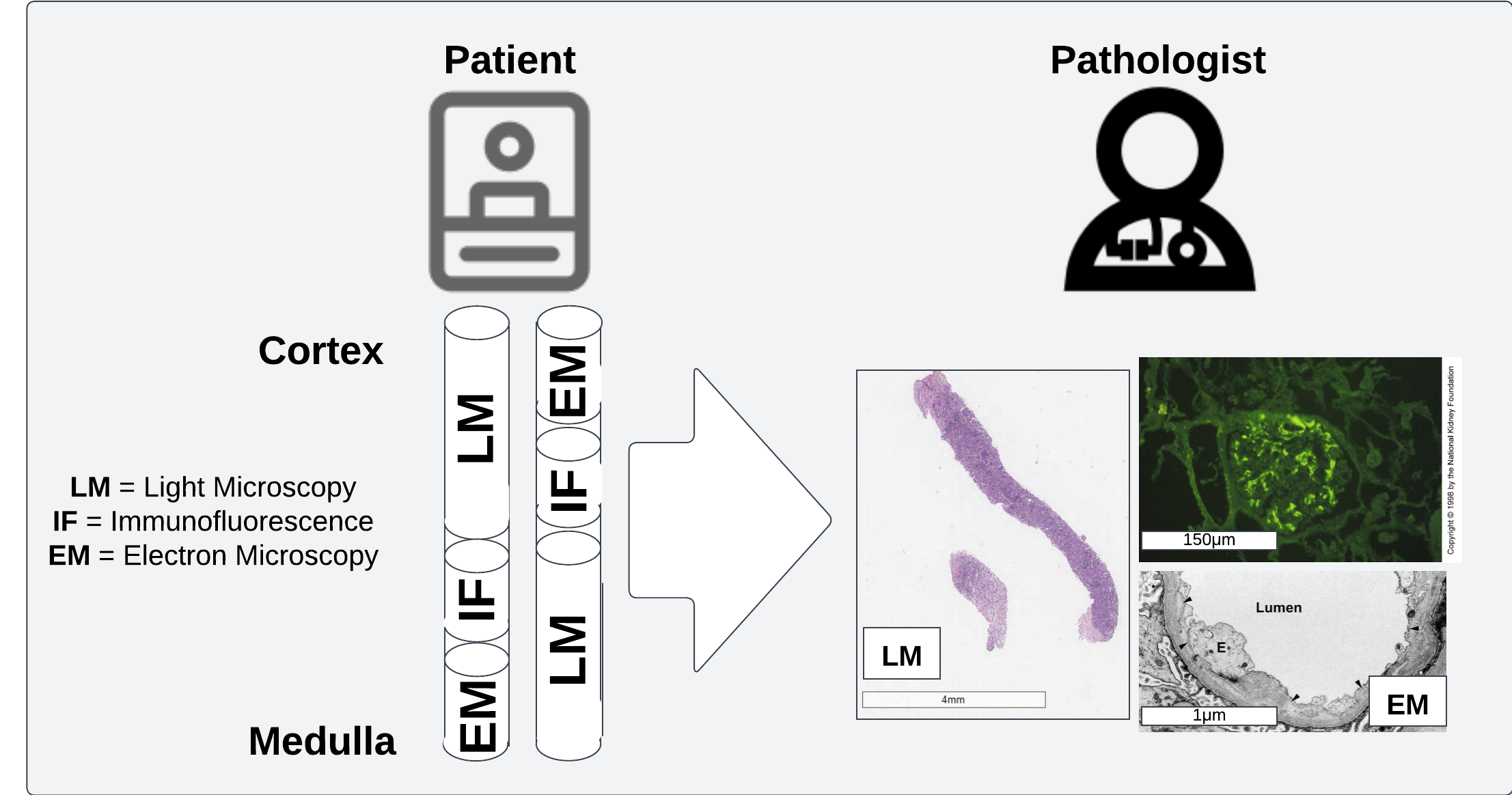 Pathologists review multiple sections from each patient with different preparation procedures to get a fuller view of patient health
Pathologists review multiple sections from each patient with different preparation procedures to get a fuller view of patient health
Histopatholoy is a sub-field of pathology wherein disease is studied in the context of microscopic examination of tissue structures. This can include a wide array of different scales from light microscopy to electron microscopy to better understand physical determinants of disease.
Quantification of histological structures
One of the challenges (of which there are several), in incorporating large-scale computational methods into a pathology workflow is the challenge of extracting precise measurements from histological structures. The first thing to consider here is the scale of the data which is present in even a modestly sized biopsy scan. Observations which can be informative for diagnoses can be made at varying scales, although for broad applicability slides are usually scanned at a high resolution (~20-40X magnification). The resulting image can then be in the neighborhood of 100k pixels by 100k pixels, putting these images firmly in gigapixel territory.
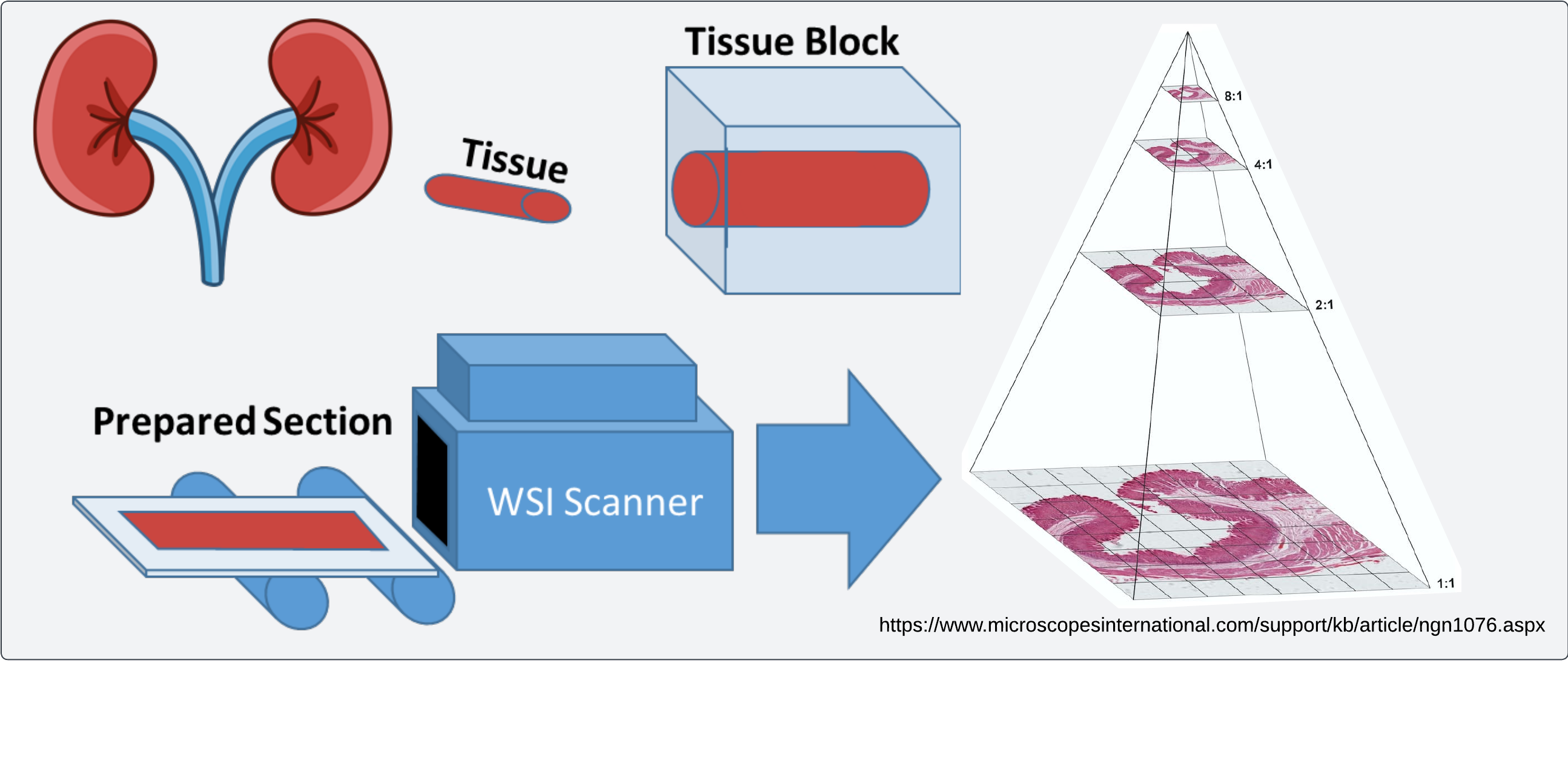 High resolution scanning of histology images enables efficient visualization and retrieval of image regions at multiple magnification levels
High resolution scanning of histology images enables efficient visualization and retrieval of image regions at multiple magnification levels
Integration of multi-modal data
Even assuming that quantification of histological structures is performed, the next challenge is integrating all available data which may include some spatial –omics method and presenting it in a way that enables generation and testing of biological hypotheses. This kind of tool would help to facilitate a special brand of analyses which require user interactions, what we call interactive analysis. This type of data analysis is especially important in pathology, where deciding what is important to look at in a slide is highly variable and qualitative. Identifying regions of interest can also require specialized knowledge which might not be shared by everyone in a team working on the same project.
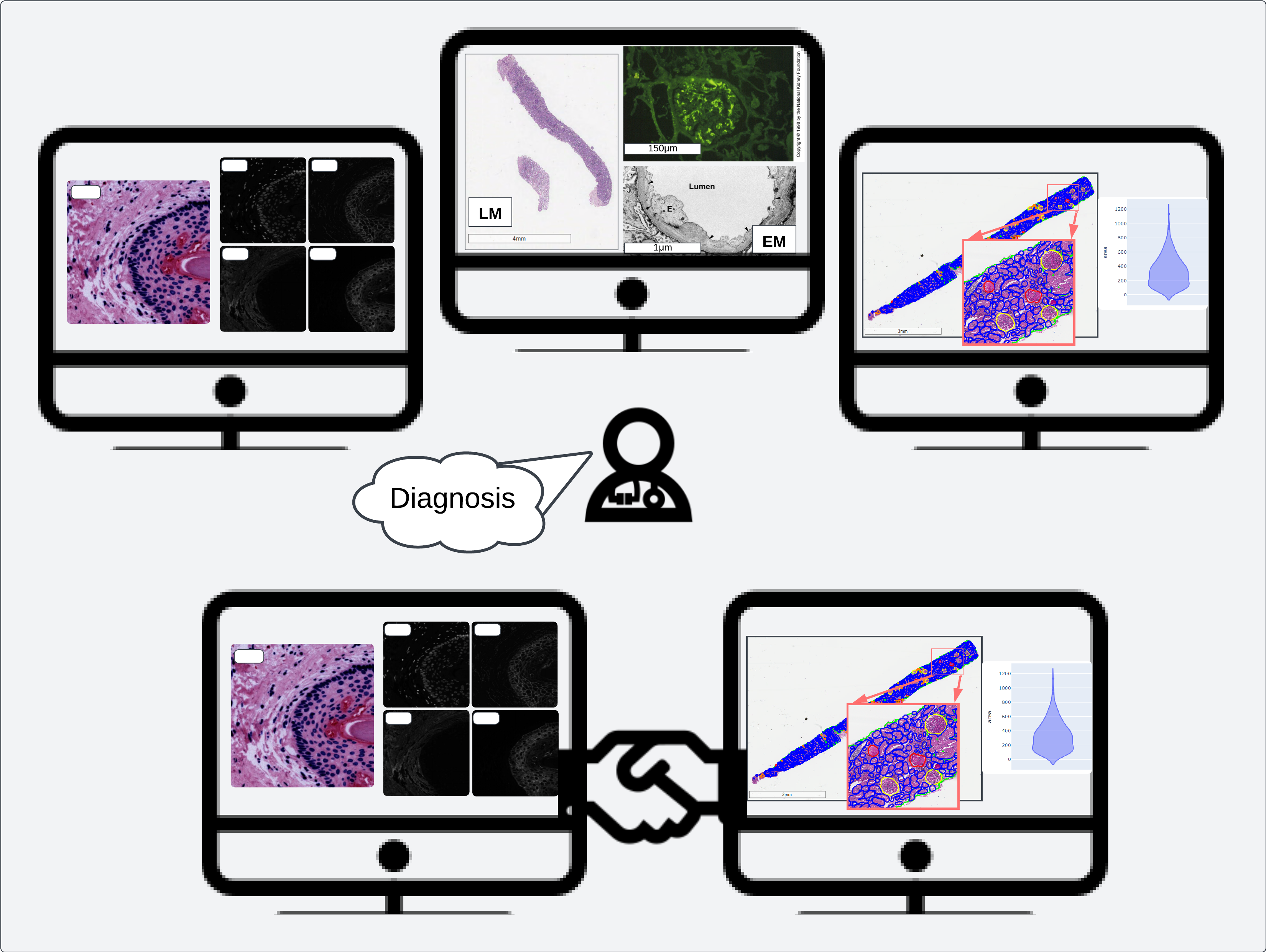 Multimodal data integration strengthens pathologist interactions with image data, empowering diagnostic and prognostic decisions with quantitative measurements and “big data”
Multimodal data integration strengthens pathologist interactions with image data, empowering diagnostic and prognostic decisions with quantitative measurements and “big data”